Publications
Highlights
(For a full list see below or go to Google Scholar)
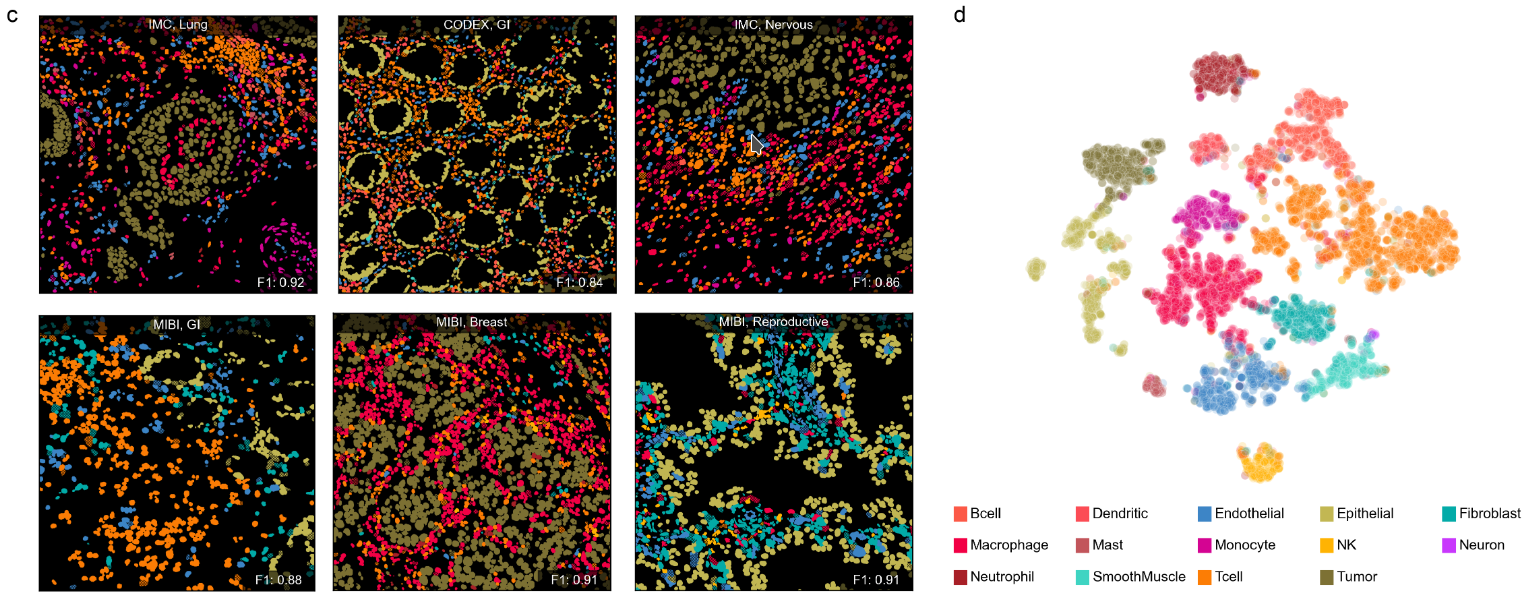
We present a novel approach to cell phenotyping for spatial proteomics that addresses the challenge of generalization across diverse datasets with varying marker panels. Our approach utilizes a transformer with channel-wise attention to create a language-informed vision model; this model’s semantic understanding of the underlying marker panel enables it to learn from and adapt to heterogeneous datasets. Leveraging a curated, diverse dataset with cell type labels spanning the literature and the NIH Human BioMolecular Atlas Program (HuBMAP) consortium, our model demonstrates robust performance across various cell types, tissues, and imaging modalities. Comprehensive benchmarking shows superior accuracy and generalizability of our method compared to existing methods. This work significantly advances automated spatial proteomics analysis, offering a generalizable and scalable solution for cell phenotyping that meets the demands of multiplexed imaging data.
X Wang*, R Dilip, Y Bussi, C Brown, E Pradhan, Y Jain, K Yu, M Abt, K Börner, L Keren, Y Yue, R Barnowski, D Van Valen*
(Preprint) bioaRxiv-10.1101/2024.11.02.621624
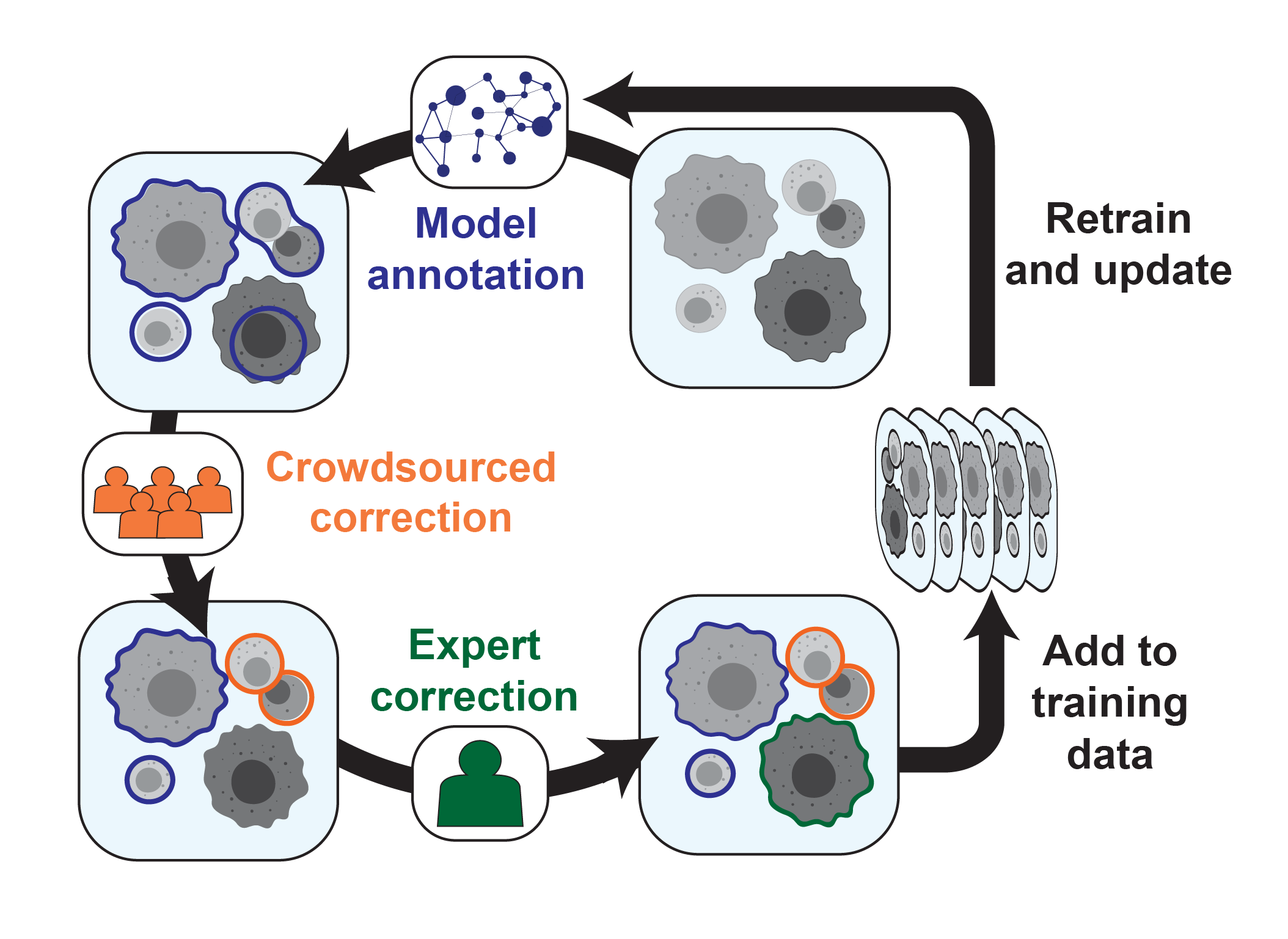
Cell segmentation is a significant challenge for tissue imaging experiments. While deep learning offers a promising solution, existing methods are limited by a lack of training data. Here we used crowdsourcing and a human-in-the-loop approach to annotate over 1 million cell nuclei and whole cells in a diverse array of tissue images. We used the resulting dataset to train deep learning models that perform whole-cell segmentation with human level performance.
N Greenwald*, G Miller*, E Moen, A Kong, A Kagel, C Fullaway, B McIntosh, K Leow, M Schwartz, T Dougherty, C Pavelchek, S Cui, I Camplisson, O Bar-Tal, J Singh, M Fong, G Chaudhry, Z Abraham, J Moseley, S Warshawsky, E Soon, S Greenbaum, T Risom, T Hollmann, L Keren, W Graf, M Angelo*, and D Van Valen*
Nature Biotechnology 10.1038/s41587-021-01094-0
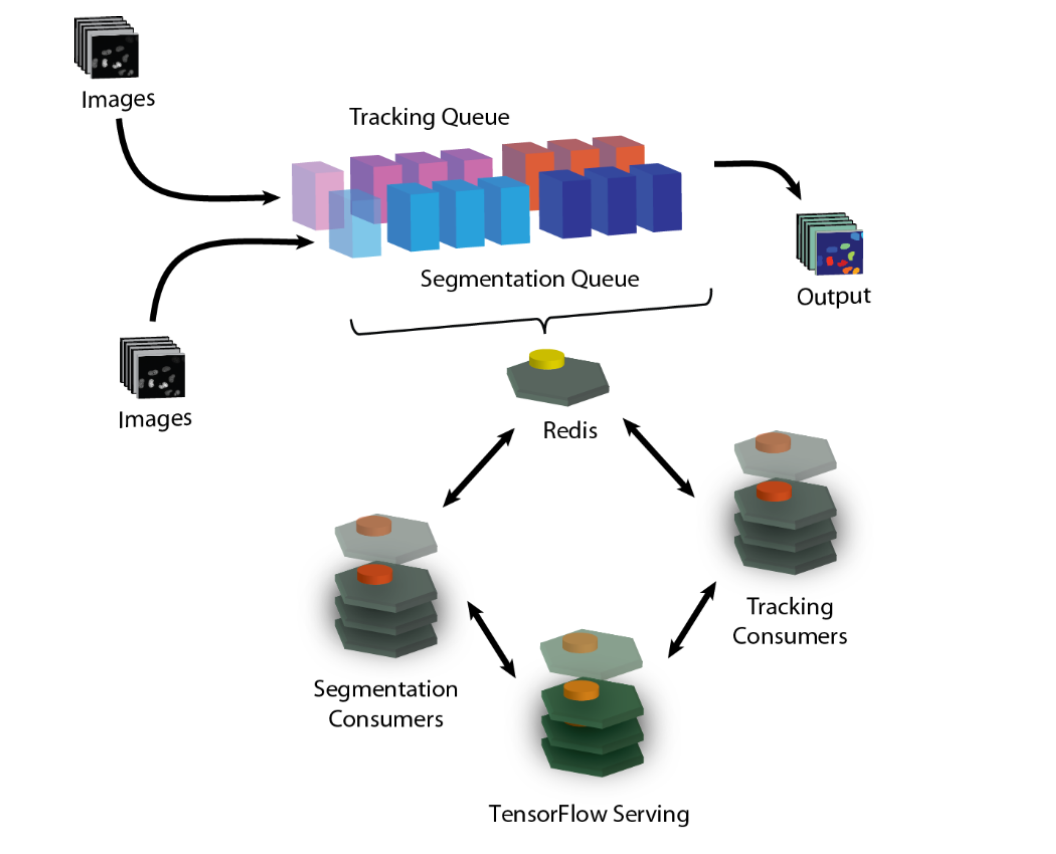
Deep learning is transforming the analysis of biological images, but applying these models to large datasets remains challenging. Here we describe the DeepCell Kiosk, cloud-native software that dynamically scales deep learning workflows to accommodate large imaging datasets. To demonstrate the scalability and affordability of this software, we identified cell nuclei in 106 1-megapixel images in ~5.5 h for ~US$250, with a cost below US$100 achievable depending on cluster configuration.
D Bannon, E Moen, M Schwartz, E Borba, T Kudo, N Greenwald, V Vijayakumar, B Chang, E Pao, E Osterman, W Graf and D Van Valen
Nature Methods (18), 43-45 (2021)

Single cell analysis of live-cell imaging experiments requires identifying cells in single frames and linking them together over time. In this work, we solve the segmentation and linkage problems by creating a large annotated dataset for live-cell imaging. We use this data to train deep learning models that achieve state-of-the-art performance in cell tracking and lineage construction.
E Moen, E Borba, G Miller, M Schwartz, D Bannon, N Koe, I Camplisson, D Kyme, C Pavelchek, T Price, T Kudo, E Pao, W Graf, and D Van Valen
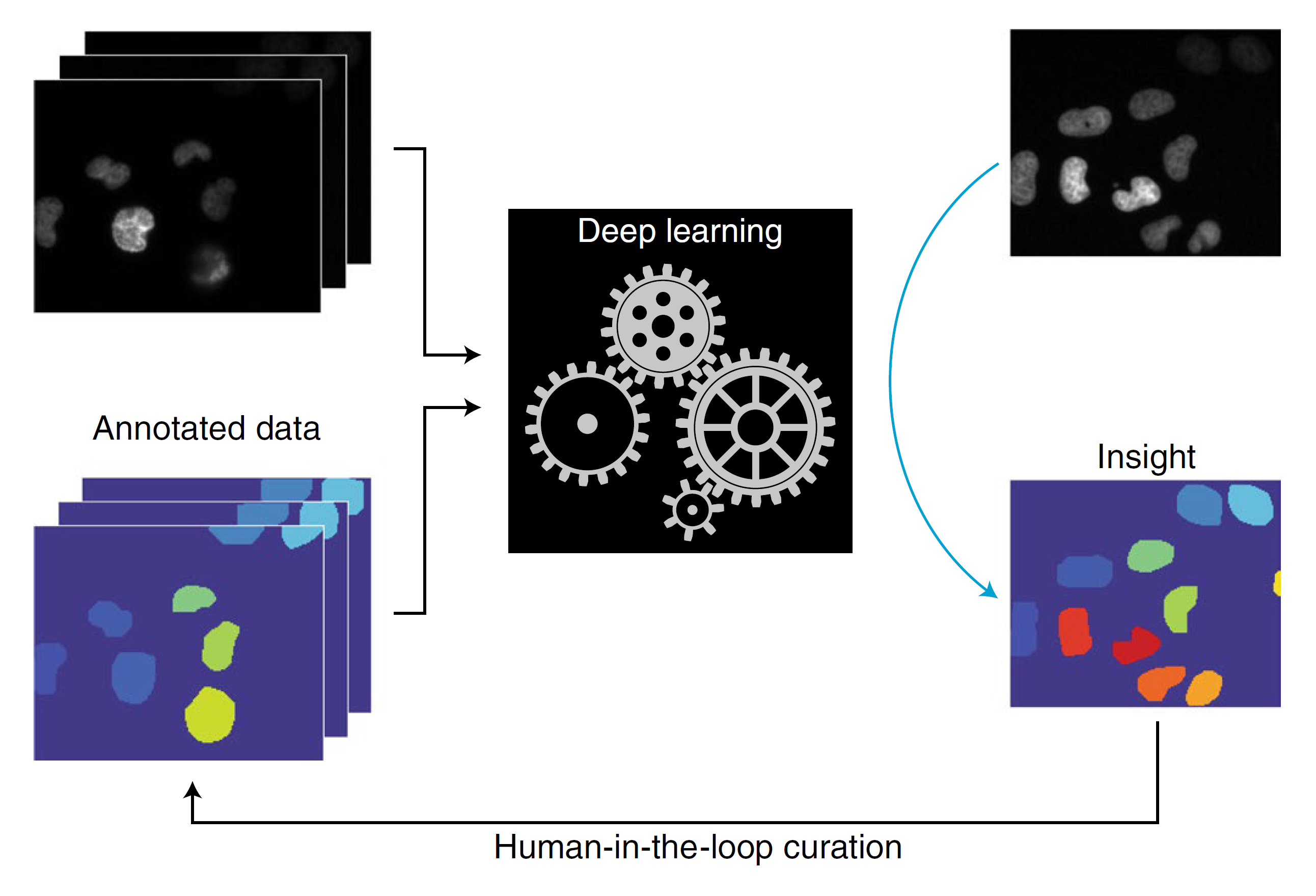
Deep learning is transforming our ability to analyze the behavior of cells in microscope images. In this review, we survey the field’s progress on adapting these methods to perform classification, segmentation, object tracking, and latent information extraction on biological images. We point readers to existing software packages for each application, as well as to annotated datasets.
E Moen, D Bannon, T Kudo, W Graf, M Covert, and D Van Valen
Nature Methods (1), 1-14 (2019)
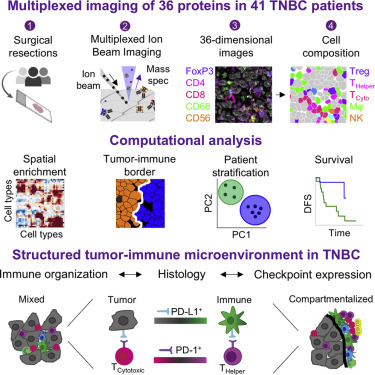
Spatial genomics technologies let us measure “omics” type data in a fashion that preserves spatial information, but the data analysis is challenging. We partnered with the Angelo lab to apply DeepCell to spatial proteomics data of triple-negative breast cancer to quantify the interactions between immune cells and tumor cells in the tumor microenvironment.
L Keren, M Bosse, D Marquez, R Angoshtari, S Jain, S Varma, S Yang, A Kurian, D Van Valen, R West, S Bendall, and M Angelo
Cell 174 (6), 1373-1387. e19 (2018).
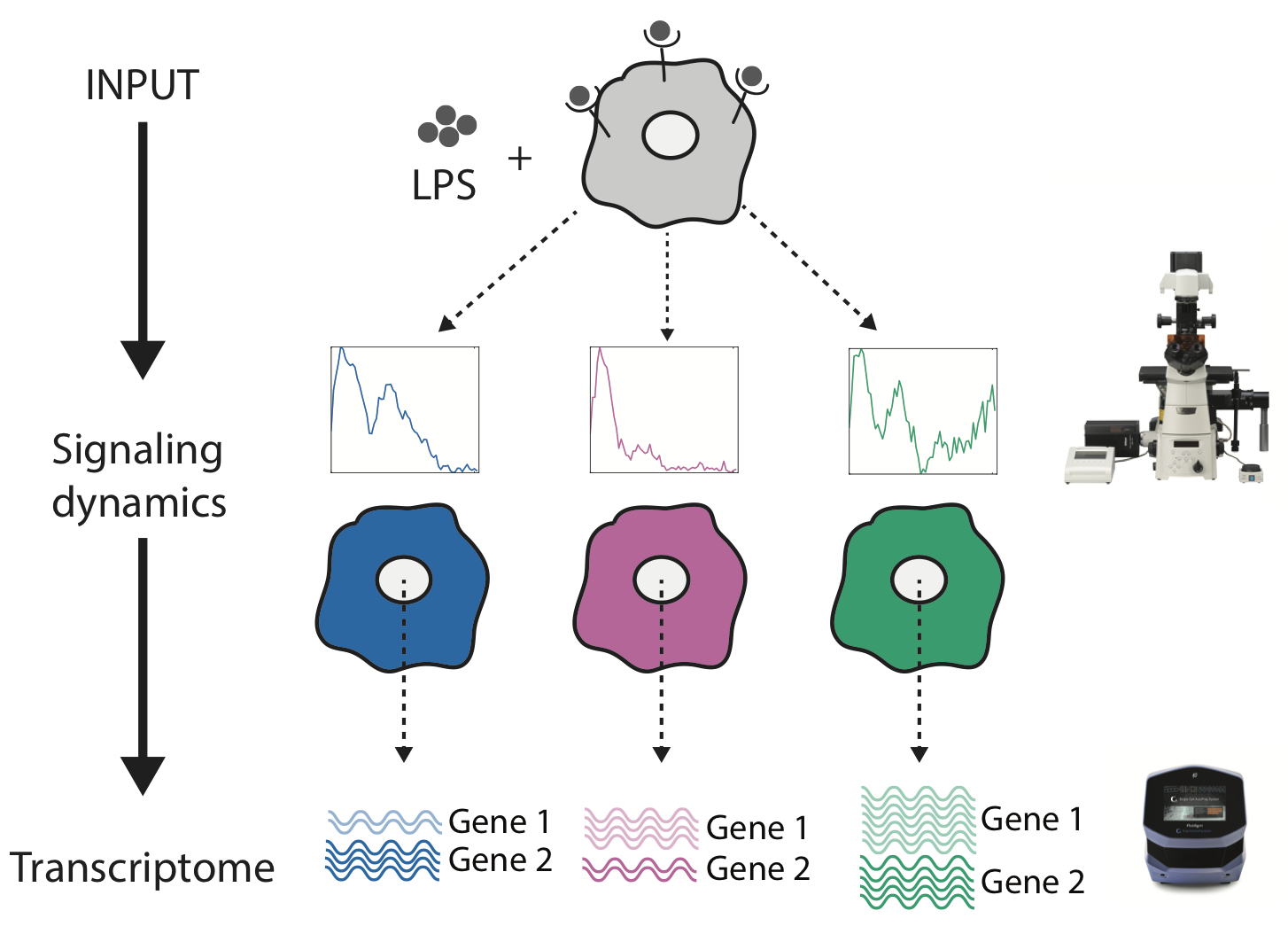
Understanding how signaling dynamics are decoded into patterns of gene expression is challenging because imaging and genomics technologies don’t talk to each other. In this work, we performed live-cell imaging and single-cell RNA sequencing in the same individual cell to understand how NF-κB dynamics are turned into distinct patterns of gene expression.
K Lane*, D Van Valen*, M DeFelice, D Macklin, T Kudo, A Jaimovich, A Carr, T Meyer, D Pe’er, S Boutet, and M Covert
Cell Systems 4 (4), 458-469. e5 (2017).
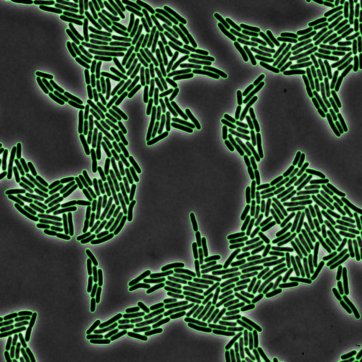
Using computer vision to obtain single-cell resolution is often the most challenging aspect of live-cell imaging experiments. In this work, we used deep learning to perform single-cell image segmentation of microscopy images across the domains of life.
D Van Valen, T Kudo, K Lane, D Macklin, N Quach, M DeFelice, I Maayan, Yu Tanouchi, E Ashley, and M Covert
PLOS Computational Biology 12 (11), 1-24 (2016).
This work was highlighted in press releases by the Hertz Foundation and CrowdFlower
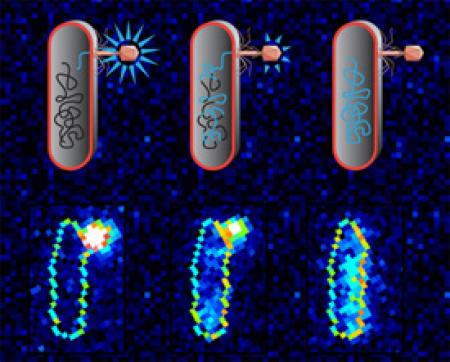
How bacteriophage lambda delivers its genome into bacterial cells is an open problem. To test different proposed biophysical mechanisms, we measured the DNA transfer between individual viruses and bacteria.
D Van Valen*, D Wu*, Y Chen, H Tuson, P Wiggins, and R Phillips
Current Biology 22 (14), 1339-1343 (2012).
This work was featured in Science magazine’s editors choice section and in a press release by Caltech.
Full List
Generalized cell phenotyping for spatial proteomics with language-informed vision models
X Wang*, R Dilip, Y Bussi, C Brown, E Pradhan, Y Jain, K Yu, M Abt, K Börner, L Keren, Y Yue, R Barnowski, D Van Valen*
(Preprint) bioaRxiv-10.1101/2024.11.02.621624
Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning
N Greenwald*, G Miller*, E Moen, A Kong, A Kagel, C Fullaway, B McIntosh, K Leow, M Schwartz, T Dougherty, C Pavelchek, S Cui, I Camplisson, O Bar-Tal, J Singh, M Fong, G Chaudhry, Z Abraham, J Moseley, S Warshawsky, E Soon, S Greenbaum, T Risom, T Hollmann, L Keren, W Graf, M Angelo*, and D Van Valen*
Nature Biotechnology 10.1038/s41587-021-01094-0
DeepCell Kiosk: scaling deep learning–enabled cellular image analysis with Kubernetes
D Bannon, E Moen, M Schwartz, E Borba, T Kudo, N Greenwald, V Vijayakumar, B Chang, E Pao, E Osterman, W Graf and D Van Valen
Nature Methods (18), 43-45 (2021)
Accurate cell tracking and lineage construction in live-cell imaging experiments with deep learning
E Moen, E Borba, G Miller, M Schwartz, D Bannon, N Koe, I Camplisson, D Kyme, C Pavelchek, T Price, T Kudo, E Pao, W Graf, and D Van Valen
bioRxiv 10.1101/803205v2
Deep learning for cellular image analysis
E Moen, D Bannon, T Kudo, W Graf, M Covert, and D Van Valen
Nature Methods (1), 1-14 (2019)
A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging
L Keren, M Bosse, D Marquez, R Angoshtari, S Jain, S Varma, S Yang, A Kurian, D Van Valen, R West, S Bendall, and M Angelo
Cell 174 (6), 1373-1387. e19 (2018).
Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-κB Activation
K Lane*, D Van Valen*, M DeFelice, D Macklin, T Kudo, A Jaimovich, A Carr, T Meyer, D Pe’er, S Boutet, and M Covert
Cell Systems 4 (4), 458-469. e5 (2017).
Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments
D Van Valen, T Kudo, K Lane, D Macklin, N Quach, M DeFelice, I Maayan, Yu Tanouchi, E Ashley, and M Covert
PLOS Computational Biology 12 (11), 1-24 (2016).
A Single-Molecule Hershey Chase Experiment
D Van Valen*, D Wu*, Y Chen, H Tuson, P Wiggins, and R Phillips
Current Biology 22 (14), 1339-1343 (2012).
Ion-dependent dynamics of DNA ejections for bacteriophage λ
D Wu*, D Van Valen*, Q Hu, and R Phillips
Biophysical journal 99 (4), 1101-1109 (2010).
Biochemistry on a leash: the roles of tether length and geometry in signal integration proteins
D Van Valen, M Haataja, and R Phillips
Biophysical journal 96 (4), 1275-1292.
*: Both authors contributed equally.